This blog is part of a broader series on drug resistance and its increasing threat to global health.
For policymakers, prioritization of vaccines for routine immunization programs is a critical exercise. Local disease burden, vaccine characteristics, and health systems capacity must all be weighed in decisions about new vaccine introductions. For some bacterial pathogens, including Salmonella Typhi (S. Typhi), which causes typhoid fever, increasing drug resistance has led to virtually untreatable infections in some parts of the world, making preventative interventions such as vaccines a vital tool for protecting communities from disease. To make the best strategic decisions for their constituents, policymakers rely on data regarding disease incidence, drug resistance, and transmission dynamics to inform decisions about where, when, and how to optimally introduce new vaccines.
Typhoid is notoriously difficult to accurately diagnose, leading to chronic underestimation of the total number of infections. While our estimates of typhoid incidence have improved significantly in recent years due to large investments from international funders, significant knowledge gaps remain. Disease burden estimates are based largely on standardized, population-based blood culture surveillance studies. Such studies have been focused largely in South Asia and more recently in sub-Saharan Africa, with relatively few data points from other parts of the world. Additionally, our global understanding of drug resistance is largely based on phenotypic tests conducted in individual laboratories.
Whole genome sequencing (WGS), or decoding the entire DNA sequence of an organism, such as S. Typhi, is easy to standardize, as genetic code is universal. The resulting genome sequence not only identifies a specific bacterial organism and describes its drug resistance profile, but also may illustrate how an individual strain has spread internationally. WGS data can also help identify the emergence of new strains that may have increased potential for treatment . When combined with local, more traditional epidemiological data, WGS data can also provide key information for the prioritization and precise targeting of public health interventions.
In a series of recently published maps, historic and contemporary data have been visualized to highlight the evolution and implications of drug-resistant typhoid. The maps use publicly available genomic data to highlight where drug-resistant typhoid is already present, but also to show the geographic spread of these strains over time. The maps highlight that drug-resistant typhoid bacteria represent a real global threat, even for countries that may not yet face a high burden, as drug-resistant organisms are regularly transferred regionally and intercontinentally.
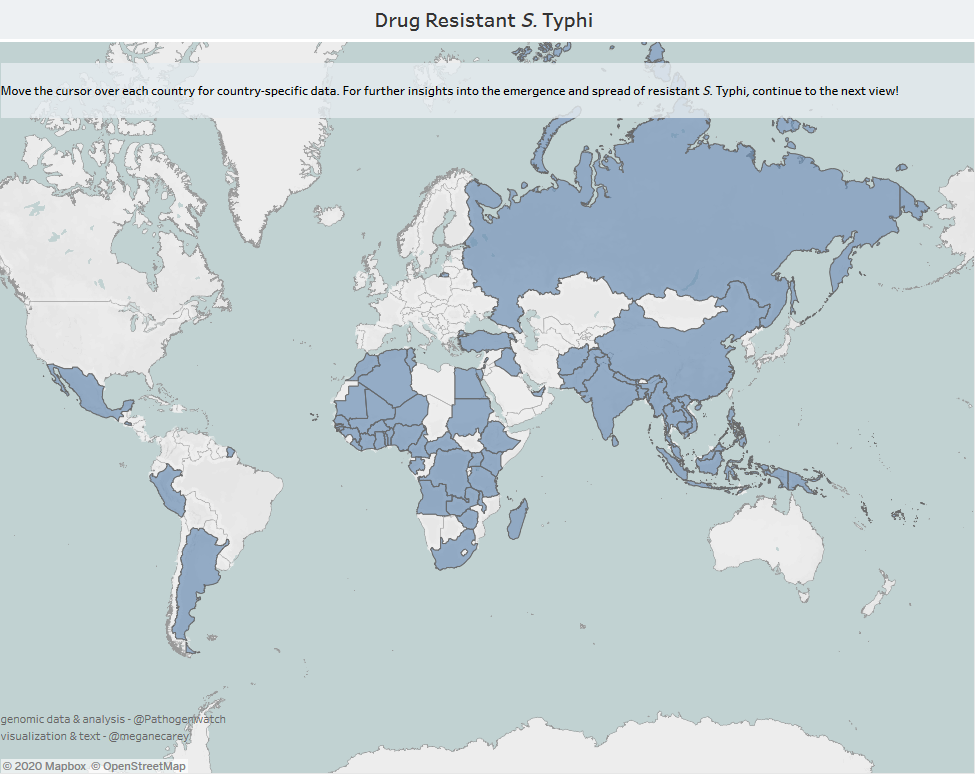
As with any dataset, there are some limitations. There are inevitably more data available from countries where funded research projects have taken place, and more data are available from recent years, as sequencing technology has improved and WGS has become more affordable. However, in spite of these limitations, the information tells an important story about the evolution of drug resistance and how it has spread during the past few decades. These graphical observations show us that drug-resistant typhoid is evolving quickly, and we are coming dangerously close to infectious organisms that are resistant to all currently available antibiotics.
With continued surveillance and research, we will continue to learn more about drug-resistant typhoid. Similarly, as more decision-makers take action to combat drug-resistant typhoid by introducing typhoid conjugate vaccines, we will learn more about how drug-resistant typhoid may be prevented. Current data show that as drug resistance continues to spread, vaccines will play an important role in reducing the impact of these organisms. While we have the data to make strategic decisions now, additional data aids in monitoring trends, allowing us to focus efforts where drug resistance continues to be a problem, and identifying any new drug-resistant typhoid strains.
We are always seeking new collaborations to improve our understanding of the molecular epidemiology of S. Typhi. If you are interested in learning more or getting involved, please contact us at typhigenomes@gmail.com.
Check out the prior blog in this series Check out the next blog in this series
Photo courtesy of Gates Archive/Samantha Reinders



